Working on digital “stuff” in the archives is always fascinating, because it blurs the borders between digital and physical. Most of the work the takes up my time is at these borders. Physical stuff requires lots of human touches to transition to “digital,” and digital stuff similarly requires lots tending by humans to ensure that it is preserved physically. After all, the 1s and 0s are stored physically somewhere, even if on the cloud or in DNA.
We’re currently working on several projects to convert physical materials to digital text. The huge quantities of rich and complicated textual material in archival collections is full of potential for use as data in both computational health research and also digital medical humanities work, but to be usable for these kinds of projects it needs to be converted to digital text or data, so that it can be interpreted by computers. To get to this point the documents must be scanned, and the scanned documents must either be transcribed, which can be immensely labor intensive, or converted directly by computers using a software that can perform Optical Character Recognition, or OCR. One of our projects using OCR to extract text from a document provides a fascinating look into the world of computer vision.
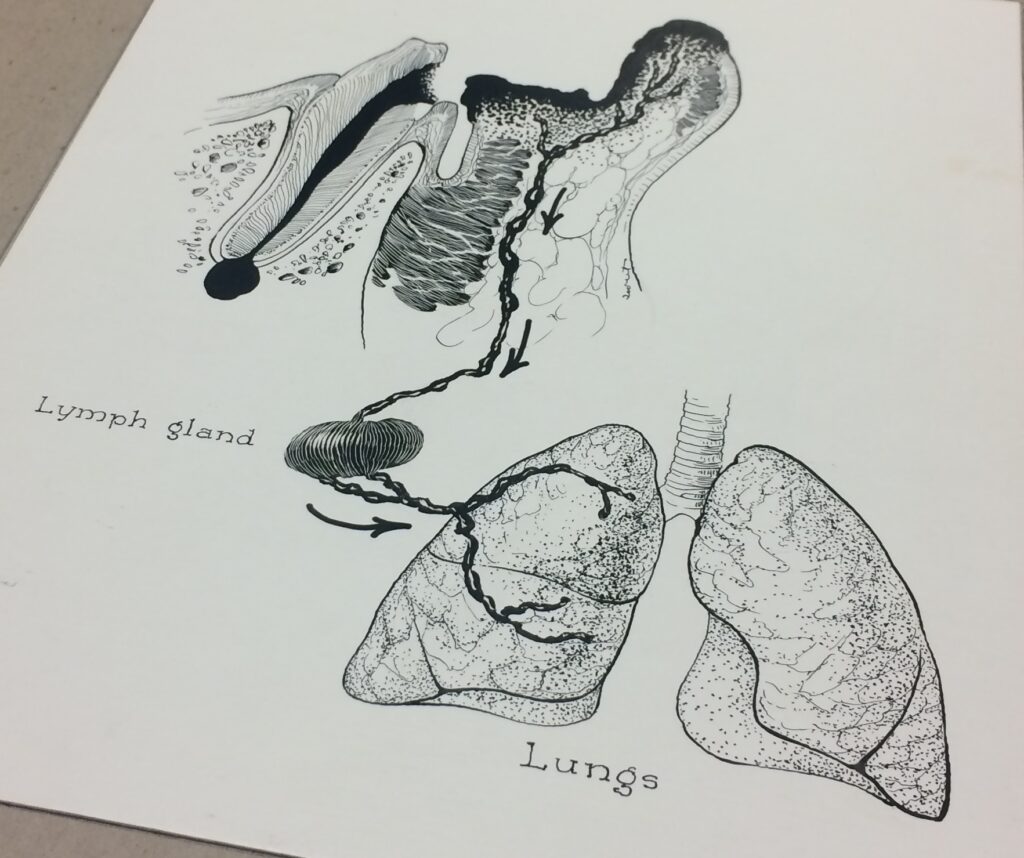
An example of the illustrations in the Ralph Sweet Collection
The Ralph Sweet Collection of Medical Illustration contains extraordinary examples of the work of one of the most renowned medical illustrators in the United States, so we’re working on digitizing the collection and putting it online. To do this we need to have detailed metadata — the kind of information you might expect to find in a catalog record, title, date, author — about each illustration. Currently this metadata for the Sweet Collection exists only in the form of printed index that was written on a typewriter. We can scan the index, but we do not have the labor to transcribe each of the 2500 or so entries. This is a job for OCR.
The image below shows what a page of the Ralph Sweet index looks like. This is the metadata that we want to be able to extract and turn into digital text so that it can be understood by a computer and used as data.

A page of the index for the Ralph Sweet Collection.
One of the first problems we encountered in attempting to extract text from this document is a classic difficulty of computer vision. As English-speaking humans, we know by looking at this document that it contains three columns, and that the order in which to read the page is top to bottom by column, starting on the left and moving right. To a computer however, it is simply a page full of text, and there is no way to know whether or not the text is broken into columns or whether each line should be read all the way across the page. This simple task presented a difficulty for most of the software that we tested, but we found one software which could identify these columns easily. The software is called Tesseract, and it was actually developed in the 1980’s but continues to be a good open-source tool to perform OCR.
If we plug the above page into Tesseract, we get mostly recognizable text, which in itself is pretty miraculous when you think about it. Looking at the text though, it quickly becomes clear that it is not an exact transcription of what’s on the page. There are misspellings (“Iivev”), and some chunks of text have been separated from the entry in which they should appear (“horizontal”).
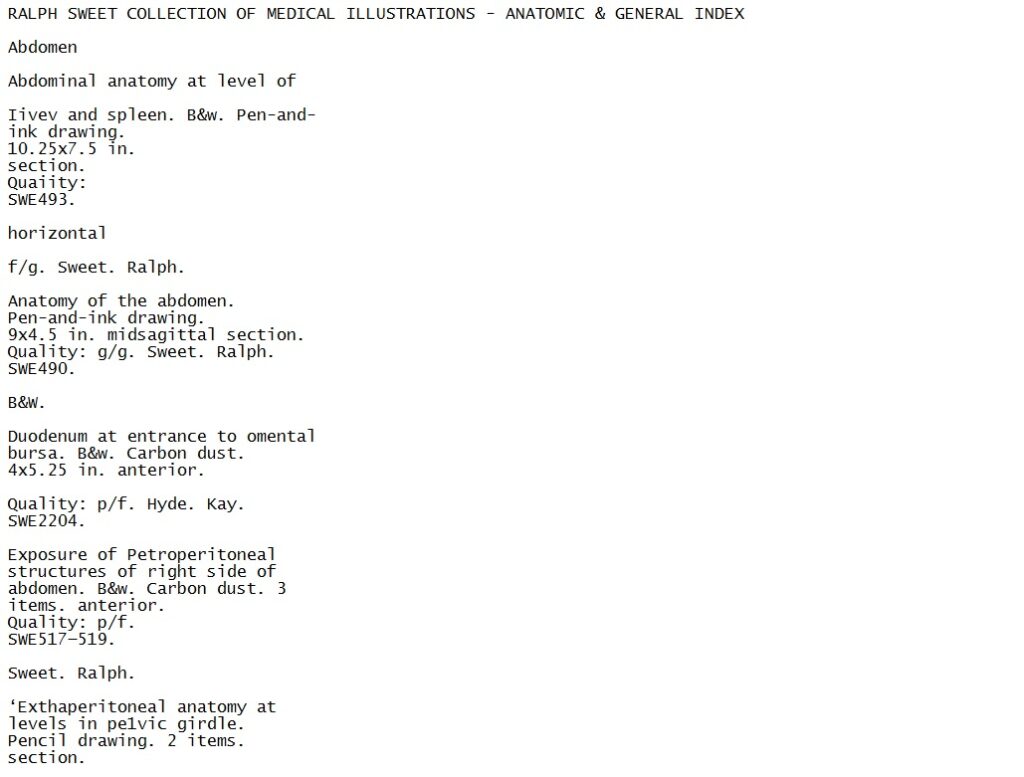
An example of the text extracted from the Ralph Sweet Collection Index by Tesseract.
Digging into the way that Tesseract (and OCR software more generally) works can help us begin to understand why these errors are cropping up. Plus, it looks really cool.
OCR programs have to go through a set of image manipulation processes to help them decide which marks on the page are text — and hence should be interpreted — and which are other marks that can be ignored. This all happen behind the scenes, and usually this involves deciding what the background parts of the image are and blurring them out, increasing the image contrast, and making the image bi-tonal so that everything on the page is only black or white. Then, the computer can trace the black pixels on the image and get a series of shapes which it can use to begin attempting to interpret as text. The image below shows the shapes that Tesseract has identified as letters and traced out for interpretation. Each new color indicates that the computer believes it has moved on to a new letter.
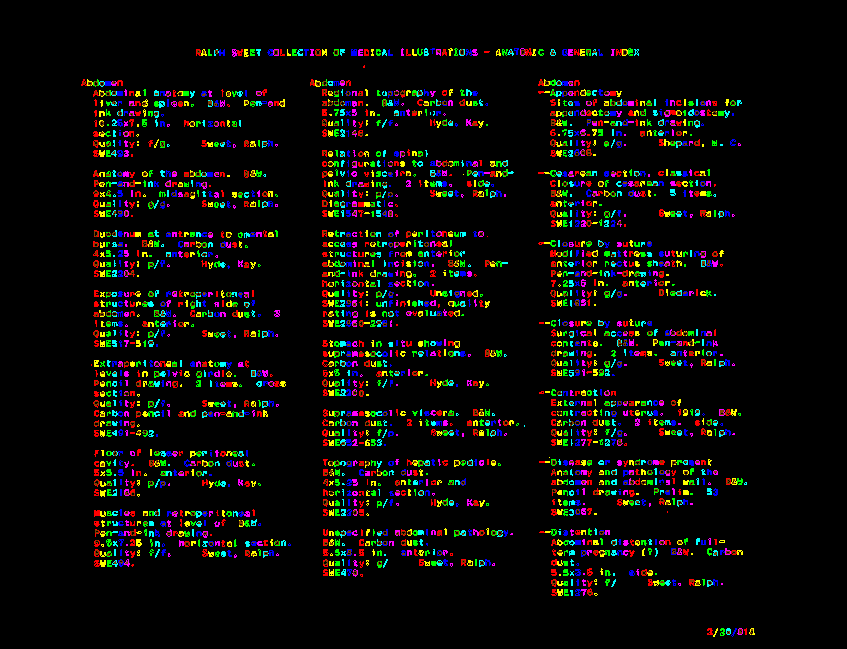
The result of Tesseract tracing the letters it has interpreted. Each new color is something that’s been identified as a new letter.
Interestingly, when comparing the computer tracing of the letters to the original image you can see that Tesseract has already made the assumption that the black spaces from the three-hole punch in the top of the page are not letters, and thus it should not bother to trace them. Lucky for us, that’s correct.
Next the computer has to take all these letters and turn them into words. In actual practice it’s not quite this simple, but basically the computer iterates on each letter identification that it believes it has made by testing whether or not that word is in its dictionary, and thus whether or not it is likely to be a word. If the combination of letters that the computer thinks it sees are not a word, then it will go back and make a new guess about the letters and test whether or not that’s a word, and so on. Part of this whole process is to chunk the letters into words using their actual spacing on the page. Below you can see an image of how Tesseract has begun to identify words using the spaces between lines and letters.

The “words” that the OCR software has identified on the page. Each blue rectangle represents a space that Tesseract has marked as containing a word.
In addition to checking the word against the dictionary though, most OCR programs also use the context of the surrounding words to attempt to make a better guess about the text. In most cases this is helpful — if the computer has read a sentence that says “the plant requires wader” it should be a relatively easy task to decide that the last word is actually “water.” In our case though, this approach breaks down. The text we want the computer to extract in this case is not sentences, but rather (meta)data. The meaning of the language has little influence on how each individual word should be read. One of the next steps for us will be trying to figure out how to better instruct Tesseract about the importance of context in making word-identification decisions (i.e., that it’s not important).
Finally, as the OCR software interprets the text it also identifies blocks of words that it believes should be grouped together, like a paragraph. Below you can see the results of this process with Tesseract.

This view shows all of the elements of Tesseract’s word identification combined. Text has been traced in color, separate letters are contained in bounding boxes, words are contained in blue rectangles, and blocks are contained in larger bounding boxes and are numbered (though the numbers are a bit difficult to see).
A line has been drawn around each block of text, and it has been given a number indicating the order in which the computer is reading it. Here we can see the source of one of the biggest problems of the OCR-generated text from earlier. Tesseract is in-accurately excluding a lot of words from their proper blocks. In the above photo, the word “Pen” is a good example. It is a part of block 20, but it has been interpreted by the computer as it’s own block — block 21 — and has been set aside to appear in the text file after block 20. Attempting to solve this problem will be our next hurdle, and hopefully we can catch you up after we are successful.
Using OCR to extract text from digital images can be a frustrating endeavor if accuracy is a necessity, but it is also a fascinating illustration of the way computers see and make decisions. Anytime we ask computers to perform tasks that interface with humans, we will always be grappling with similar issues.