Post by Geoffrey Boushey, Head of Data Engineering, UCSF Library’s Data Science and Open Scholarship Team. He teaches and consults on programming for data pipelines with an emphasis on Python, Unix and SQL.
Teaching Approaches to Data Science
Recently, Ariel Deardorff (director of the Data Science & Open Scholarship at the UCSF Library) forwarded me a paper titled “Generative AI for Data Science 101: Coding Without Learning to Code.” In this paper, the authors described how they used GitHub Copilot, a tool for generating code with AI, to supplement a fundamentals of data science course for MBA students, most of whom had no prior coding experience. Because the instructors wanted students to use AI to generate code for analysis, but not the full analysis itself, they opted for a tool that generates code without potentially “opening the Pandora’s box too wide” with ChatGPT, a tool that might blur the line between coding and analysis.They also deliberately de-emphasized the code itself, encouraging students to focus on analytical output rather than scrutinizing the R code line by line.
This approach has some interesting parallels, along with some key differences, with the way I teach programming at the UCSF library through the “Data and Document Analysis with Python, SQL, and AI” series. These workshops are attended largely by graduate students, postdocs, research staff, and faculty (people with an exceptionally strong background in research and data science) who are looking to augment their programming, machine learning, and AI skills. These researchers don’t need me to teach them science (it turns out UCSF scientists are already pretty good at science), but they do want to learn how to leverage programming and AI developments to analyze data. In these workshops, which include introductory sessions for people who have not programmed before, I encourage participants to generate their own AI-driven code. However, I have always strongly emphasized the importance of closely any code generated for analytics or data preparation, whether pulled from online examples or created through generative AI.
The goal is to engage researchers with the creative process of writing code while also guarding against biases, inaccuracies, and unintended side effects (these are issues that can arise even in code you write yourself). Although the focus on careful examination contrasts with the approach described in the paper, it made me wonder: what if I diverged in the other direction and bypassed code altogether? If the instructors were successful in teaching MBA students to generate R code without scrutinizing it, could we skip that step entirely and perform the analysis directly in ChatGPT?
Experimental Analysis with ChatGPT
As a personal experiment, I decided to recreate an analysis from a more advanced workshop in my series, where we build a machine learning model to evaluate the impact of various factors on the likelihood of a positive COVID test using a dataset from Carbon Health. I’ve taught several iterations of this workshop, starting well before Generative AI was widely available, and have more recently started integrating GenAI-generated code into the material. But this time, I thought I’d try skipping the code entirely and see how AI could handle the analysis on its own.
I got off to a slightly rocky start with data collection. The covid clinical data repository contains a years worth of testing data split into shorter CSV files representing more limited (weekly) time periods, and I was hoping I could convince ChatGPT to infer this structure from a general link to the github repository and glob all the CSV files sequentially into a pandas dataframe (a tabular data frame format). This process of munging and merging data, while common in data engineering, can be a little complicated, as github provides both a human readable and “raw” view of CSV files. Pandas needs the raw link, which requires multiple clicks through the github web interface to access. Unfortunately, I was unsuccessful in coaxing ChatGPT into reading this structure, and eventually decided to supply github with a direct link to the raw file for one of the CSV files [8]. This worked, and ChatGPT now had a pandas dataframe with about 2,000 covid test records. Ideally, I’d do this with the full ~100k row set, but for this experiment, 2,000 records was enough.
Key Findings & Limitations
Now that I had some data loaded, I asked ChatGPT to rank the features in the dataset based on their ability to predict positive COVID test results. The AI generated a reasonably solid analysis without any need for code. ChatGPT suggested using logistic regression and produced a ranked list of features. When I followed up and asked ChatGPT to use a random forest model and calculate feature importances, it did so immediately, even offering to create a bar chart to visualize the results—no coding required.
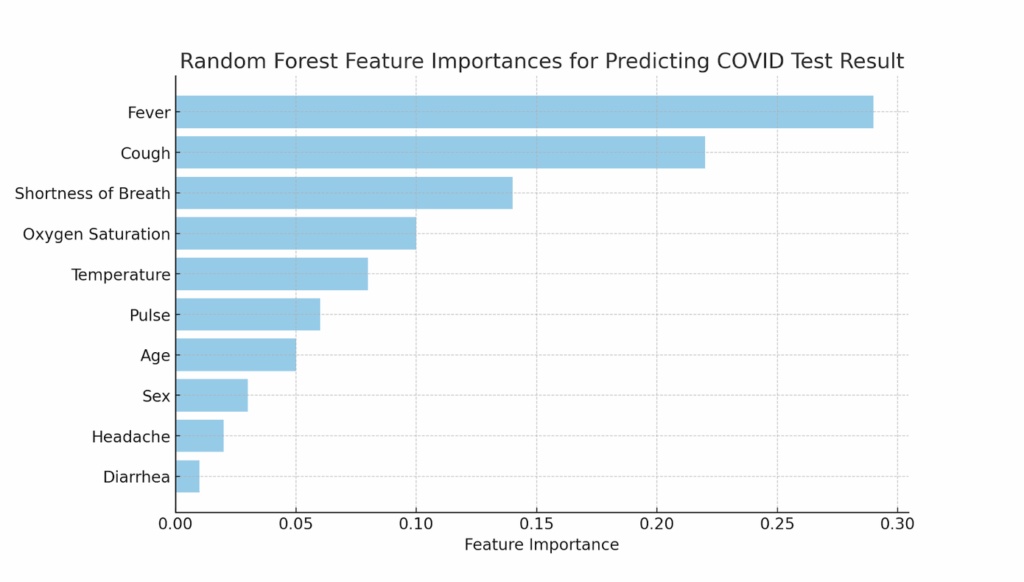
Here is the bar chart generated by ChatGPT showing the feature importances, with the inclusion of oxygen saturation but the notable omission of loss of smell:
One feature ChatGPT identified as highly significant was oxygen saturation, which I had overlooked in my prior work with the same dataset. This was a moment of insight, but there was one crucial caveat: I couldn’t validate the result in the usual way. Typically, when I generate code during a workshop, we can review it as a group and debug it to ensure that the analysis is sound. But in this no-code approach, the precise stages of this process were hidden from me. I didn’t know exactly how the model had been trained, how the data had been cleaned or missing values imputed, how the feature importances had been calculated, or whether the results had been cross-validated. I also didn’t have access to the feature importance scores from some machine learning algorithms (such as random forest) that I had built and trained myself. The insight was valuable, but it was hard to fully trust or even understand it without transparency into the process.

This lack of transparency became even more apparent when I asked ChatGPT about a feature that wasn’t showing up in the results: loss_of_smell. When I mentioned, through the chat interface, that this seems a likely predictor for a positive test and asked why it hadn’t been included, ChatGPT told me that this feature would indeed be valuable and articulated why, but repeated that it wasn’t part of the dataset. This surprised me, as it was in the dataset under the column name “loss_of_smell”
(The full transcript of this interaction, including the AI’s responses and corrections, can be found in the footnote [1] below).
This exchange illustrated both the potential and the limitations of AI-powered tools. The tool was quick, efficient, and pointed me to a feature I hadn’t considered. But it still needed human oversight. Tools like ChatGPT can miss straightforward details or introduce small errors that a person familiar with the data might easily catch. It could also introduce errors that may be notably more obscure, and only detectable after very careful examination and consideration of the output by someone with a much deeper knowledge of the data.
The Importance of Understanding Your Data
The experience reinforced a key principle I emphasize in teaching: know your data. Before jumping into analysis, it’s important to understand how your data was collected, how it’s been processed, what (ahem) a data engineering team may have done to it as they prepared it for you, and what it represents. Without that context, you may not be able to know when AI or other tools have led you in the wrong direction or missed critical context.
While the experiment I conducted with AI analysis was fascinating and demonstrates the potential of low- or no-code approaches, it does underscore, for me, the continued importance of generating and carefully reading code during my programming workshops. Machine learning tasks related to document analysis, such as classification, regression, and feature analysis, involve a very precise set of instructions for gathering, cleaning, formatting, processing, analyzing, and visualizing data. While generative AI tools often provide quick, opaque results, the precision involved in these processes can be obscured from the user. This lack of transparency carries significant implications for repeatable research and proper validation. For now, it remains crucial to have access to the underlying code and understand its workings to ensure thorough validation and review.
Conclusions & Recommendations
Programming languages will likely remain a crucial part of a data scientist’s toolkit for the foreseeable future. But whether you are generating and verifying code or using AI directly on a dataset, keep in mind that the core of data science is the data itself, not the tools used to analyze it.
No matter which tool you choose, the most important step is to deeply understand your data – its origins, how it was collected, any transformations it has undergone, and what it represents. Take the time to engage with it, ask thoughtful questions, and stay vigilant about potential biases or gaps that could influence your analysis. (Is it asking too much to suggest you love your data? Probably. But either way, you might enjoy the UC-wide “Love Data Week” conference).
In academic libraries, much of the value of archives comes from the richness of the objects themselves—features that don’t necessarily come through in a digital format. This is why I encourage researchers to not just work with digital transcriptions, but to also consider the physicality of the data: the texture of the paper, the marks or annotations on the margins, and the context behind how that data came to be. These details often carry meaning that isn’t immediately obvious in a dataset or a plain text transcription. Even in the digital realm, knowing the context, understanding how the data was collected, and remaining aware of the possibility of hidden bias are essential parts of the research process. [3] [4] Similarly, when working with archives or historical records, consider the importance of engaging with the data beyond just the text transcript or list of AI-detected objects in images.
Get to know your data, before, during, and after you analyze it. If possible, handle documents physically, consider what you may have missed. Visit the libraries, museums, and archives [12] where objects are physically stored, talk to archivists and curators who work with them. Your data will tend to outlast the technology that you use to analyze it, and while the tools and techniques you use for analysis will evolve, your knowledge of your data will form the core of its long-term value to you.
- GPT transcript: https://github.com/geoffswc/Know-Your-Data-Post/blob/main/GPT_Transcript.pdf
- In the workshop series, we use python to merge the separate csv files into a single pandas dataframe using the python glob module. It wouldn’t be difficult to do this and resume working with ChatGPT, though it does demonstrate the difficulty of completing an analysis without any manual intervention through code (for now).
- “Bias and Data Loss in Transcript Generation” UCTech, 2023: https://www.youtube.com/watch?v=sNNrx1i96wc
- Leveraging AI for Document Analysis in Archival Research and Publishing, It’s About a Billion Lives Symposium 2025 (recording to be posted) https://tobacco.ucsf.edu/it%E2%80%99s-about-billion-lives-annual-symposium
- https://www.library.ucsf.edu/archives/ucsf/